

The details for molecular weights and concentrations are provided in Table 2. d Hydrodynamic radius for frequently used molecular weight standards to calibrate size exclusion column was measured at single concentration. c Plot of R h vs concentration for ovalbulin, where the y-axis intercept provides concentration-independent R h of 2.88 ± 0.06 nm. For biophysical and structure biology applications, such preparations are acceptable however, for therapeutic antibodies, further optimization may be required. In both cases, nearly identical positions of peak suggest a homogenous preparation. However, since large molecules scatter light more strongly, even a trace amount of aggregation can be detected using intensity distribution. Those peaks are not present in volume distribution profiles ( b) since the amount of aggregated material is negligible. The intensity distributions ( a) suggest presence of ∼5 % aggregated material at 100 nm and above. a and b represents R h distribution at multiples concentrations for ovalbumin. Recombinant ND-1 (139 kDa) was expressed into 293-EBNA cells and purified using affinity chromatography, followed by SEC in 50 mM Tris–HCl (pH 7.5), 200 mM NaCl buffer as described earlier (Patel et al. d Homogeneity of ND-1 reflected by volume distribution of nearly identical peaks from multiple measurements, demonstrating highly purified and homogenous preparation. c Similar to a, the aggregation studies for ND-1 were performed at 0.5 mg/m for a preparation optimized to obtain cleaner sample, indicating that compared to a, the level of aggregation is severely reduced. Again, the degradation is visible for the measurement that is clearly visible for intensity distribution. However, peaks from multiple runs do not coincide with each other: that essentially is a reflection of heterogeneous sample. b Hydrodynamic radius distribution by volume derived from intensity profiles for ND-1 at 0.4 mg/ml concentration, suggesting that volume distribution is not sensitive enough for detecting scant amount of aggregation. Degradation at 4 nm is also visible for one of the measurements in red colour. a Hydrodynamic radius distribution by intensity for nidogen-1 (ND-1) at 0.4 mg/ml, indicating that although there is a major peak for each measurement, there are multiple peaks visible between 15 nm and 500 nm ( x-axis) suggesting the presence of high molecular weight aggregated species. 6 Alberta RNA Research and Training Institute, Department of Chemistry & Biochemistry, University of Lethbridge, 4401 University Drive, Lethbridge, Alberta, T1K 3M4, Canada. 5 School of Biosciences, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK.4 Department of Biochemistry and Medical Genetics, University of Manitoba, 745 Bannatyne Avenue - Basic Medical Sciences Building, Winnipeg, Manitoba, R3E 0J9, Canada.3 Department of Chemistry, University of Manitoba, 144 Dysart Road, Winnipeg, Manitoba, R3T 2N2, Canada.

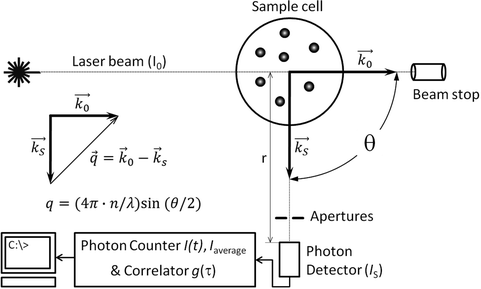
2 Department of Biochemistry and Medical Genetics, University of Manitoba, 745 Bannatyne Avenue - Basic Medical Sciences Building, Winnipeg, Manitoba, R3E 0J9, Canada. 1 Department of Chemistry, University of Manitoba, 144 Dysart Road, Winnipeg, Manitoba, R3T 2N2, Canada.


 0 kommentar(er)
0 kommentar(er)
